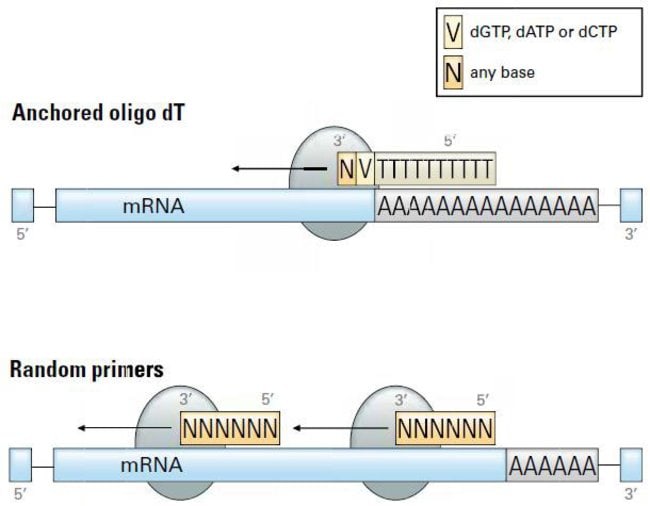
Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription | PNAS
Qualitative De Novo Analysis of Full Length cDNA and Quantitative Analysis of Gene Expression for Common Marmoset (Callithrix jacchus) Transcriptomes Using Parallel Long-Read Technology and Short-Read Sequencing | PLOS ONE
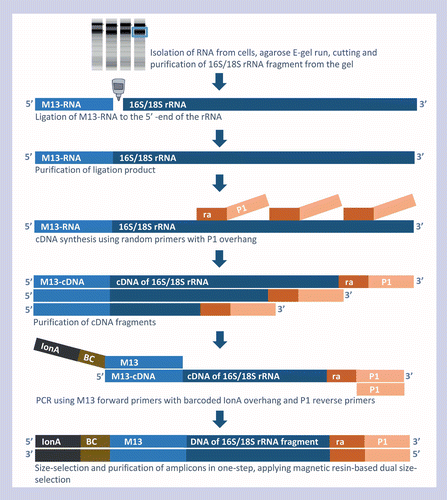
Differential priming of RNA templates during cDNA synthesis markedly affects both accuracy and reproducibility of quantitative c
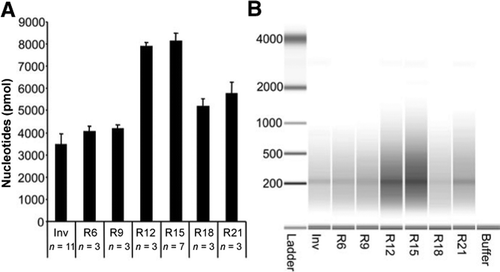
Reverse transcription using random pentadecamer primers increases yield and quality of resulting cDNA | BioTechniques

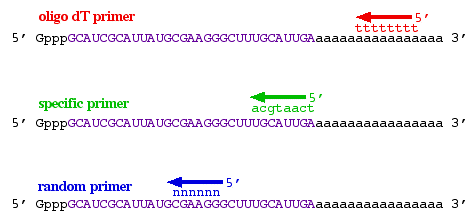
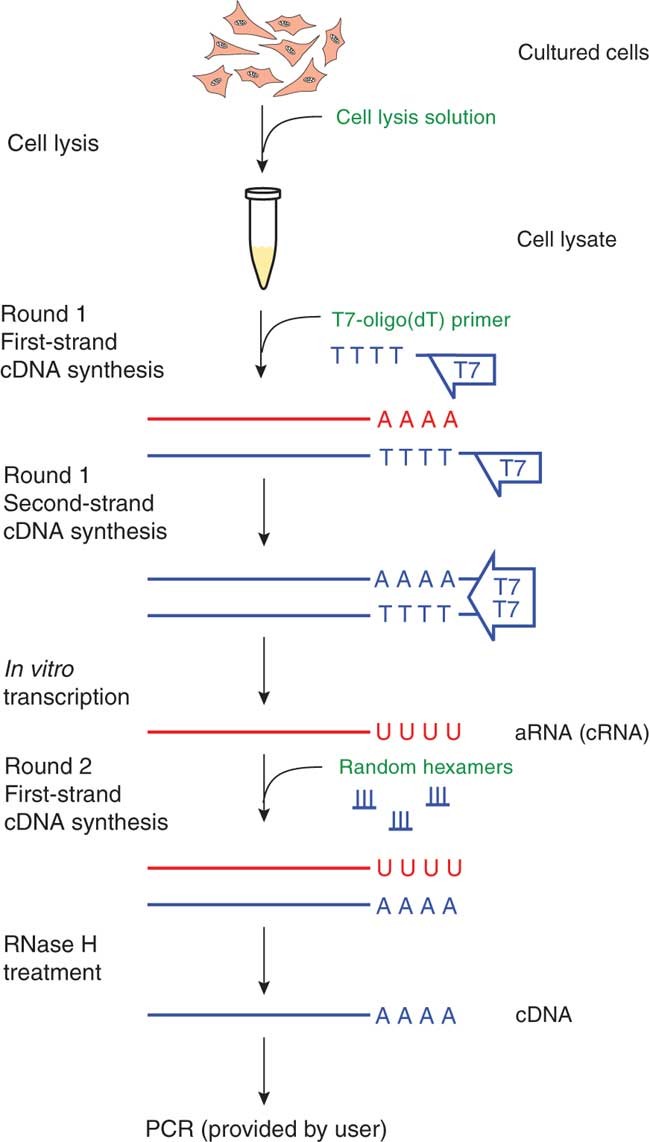
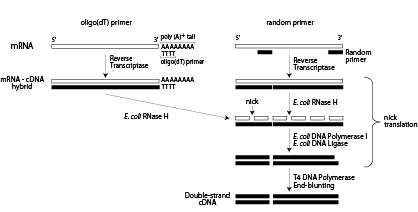
NZYTech, Genes and Enzymes - What primers are used for Reverse Transcription? There are three different approaches for priming cDNA reactions: oligo(dT) primers, random primers, or sequence-specific primers. These primers differently bind
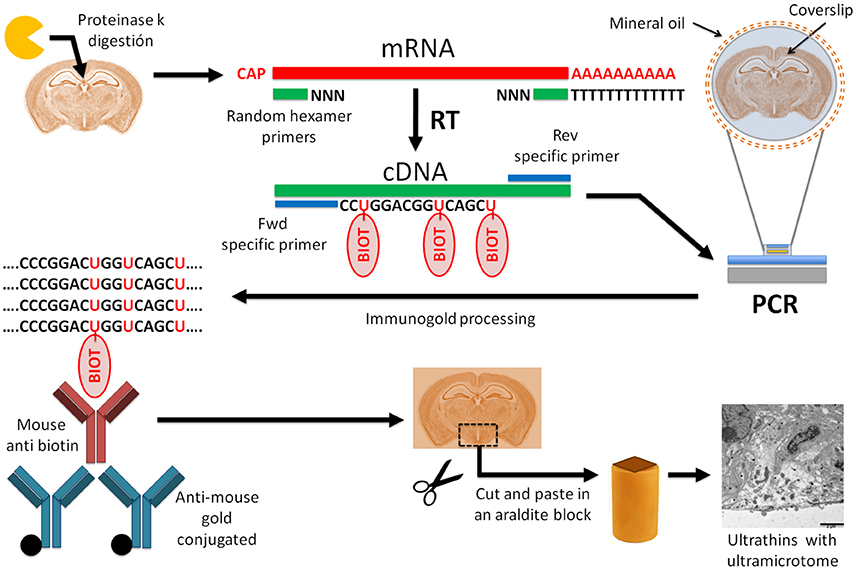

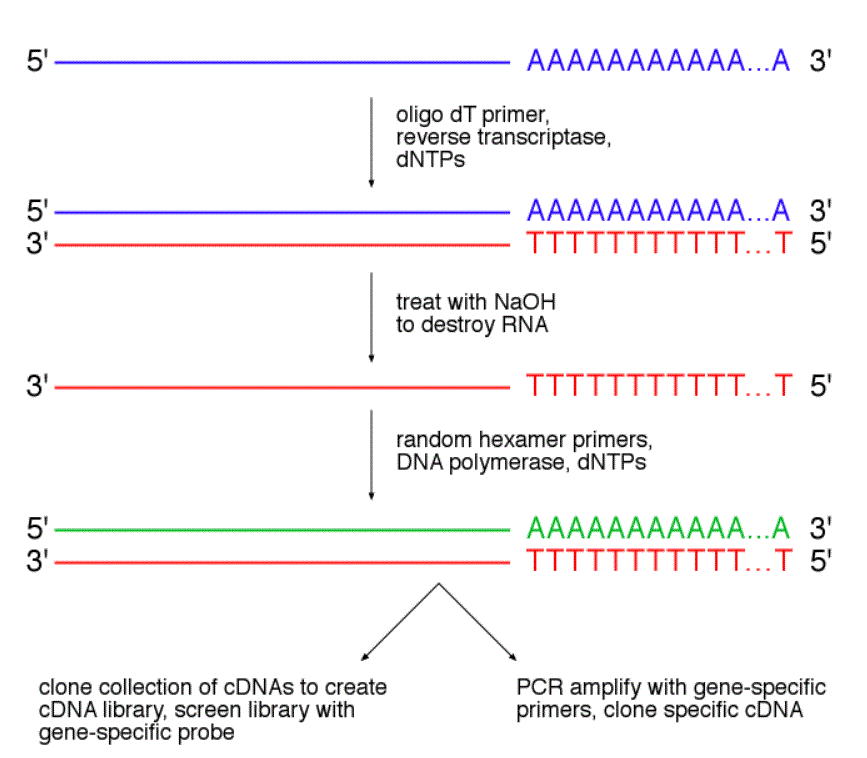







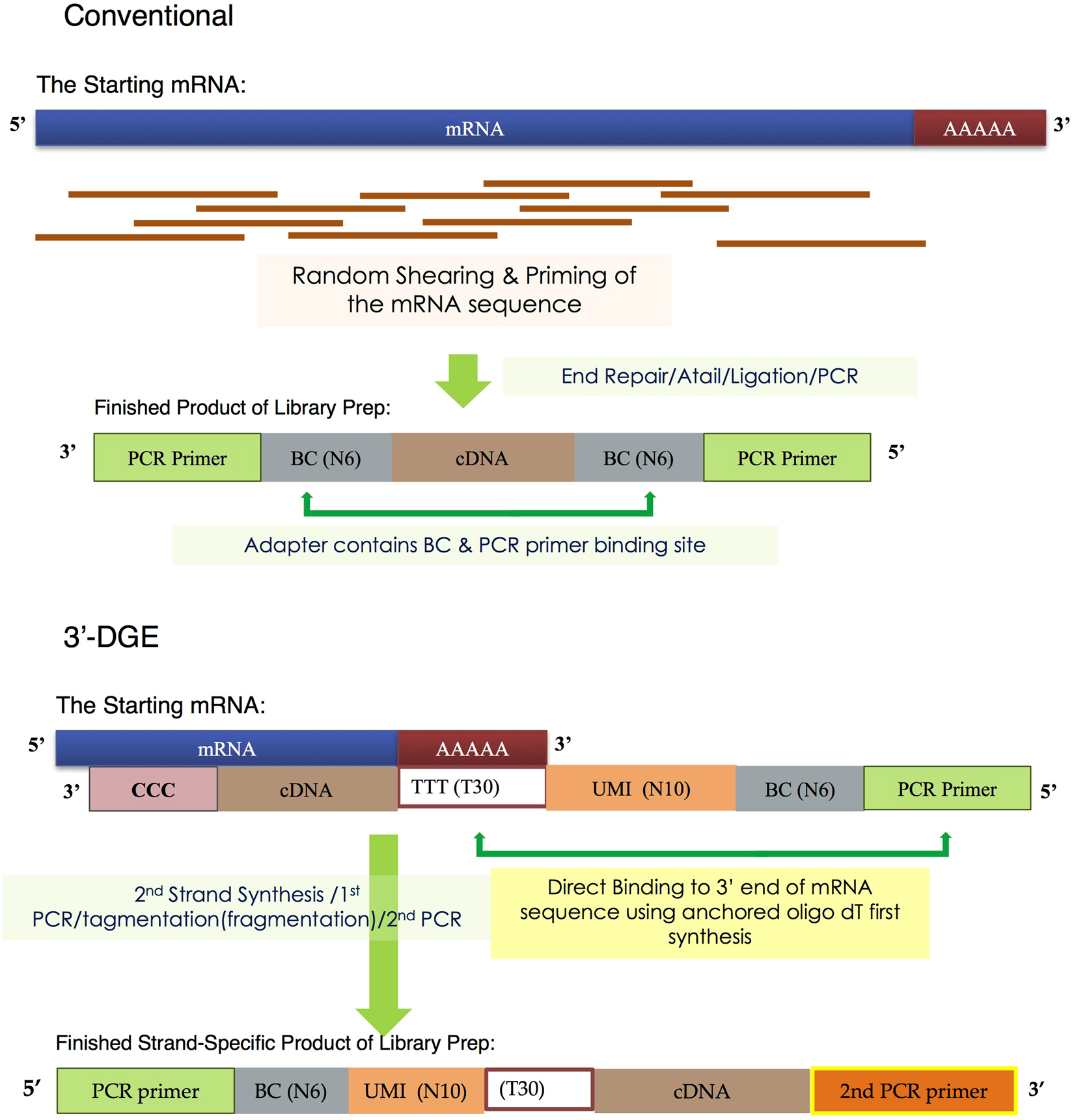

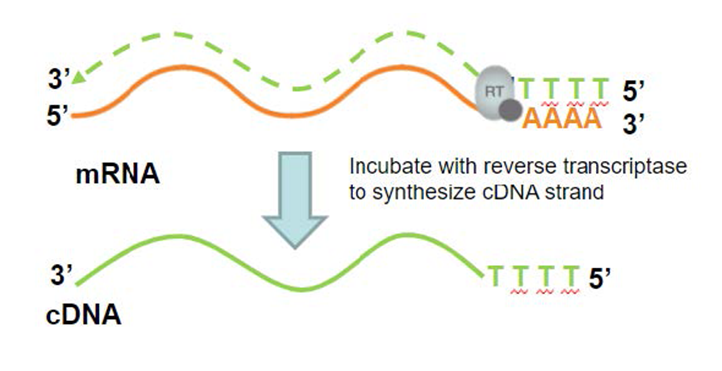
![[RP1300] ExcelRT™ Reverse Transcription Kit, 100 Rxn [RP1300] ExcelRT™ Reverse Transcription Kit, 100 Rxn](https://www.smobio.com/web/image/2311?access_token=6eb5e6fa-8d5c-4518-8212-7f5e749c8e36)